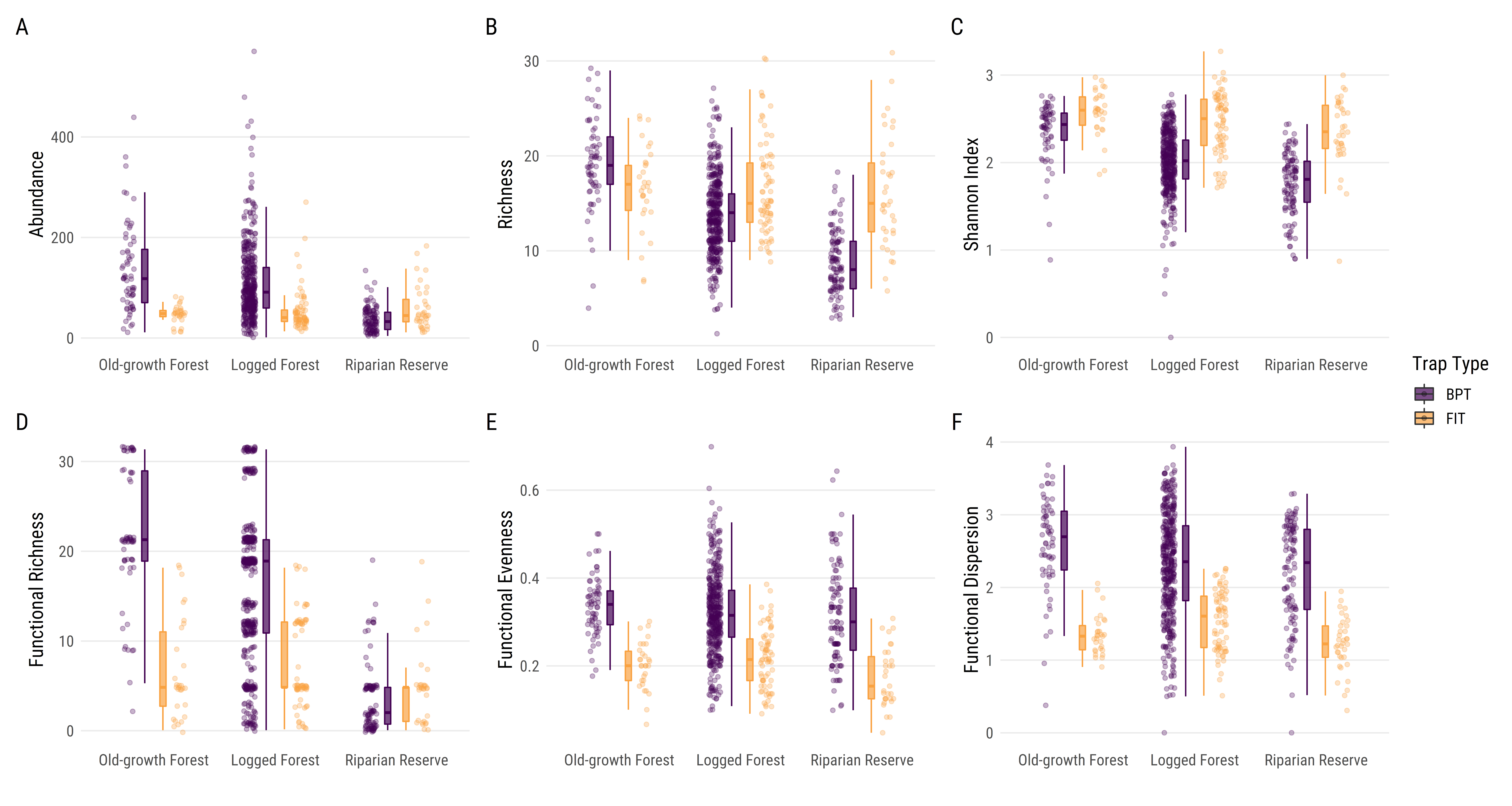
# Setup library (tidyverse)library (patchwork)windowsFonts (Roboto= windowsFont ("Roboto Condensed" ))# Read data <- read.csv ("Outputs/updated aug 2021/metrics/complete_metrics.csv" , header = T, fileEncoding= "UTF-8-BOM" )# Prepare individual plots <- ggplot (complete_metrics, aes (x = habitat, y = abundance,colour = trap, fill = trap)) + geom_boxplot (width = 0.1 , outlier.shape = NA , alpha = 0.7 , position = position_dodge (0.3 ) ) + geom_point (size = 1.3 ,alpha = .3 ,position = position_jitterdodge (dodge.width = 0.85 ,jitter.width = 0.1 )) + xlab ("" ) + ylab ("Abundance" ) + scale_colour_manual (values = c ("#440154FF" , "#F9A242FF" ),guide = "none" ) + scale_fill_manual (name = "Trap Type" ,values = c ("#440154FF" , "#F9A242FF" )) + scale_x_discrete (limits = c ("Old-growth Forest" ,"Logged Forest" ,"Riparian Reserve" )) + theme_minimal () + theme (panel.grid.minor.y = element_blank (),panel.grid.major.x = element_blank (),text = element_text (family = "Roboto" ,size = 14 ))<- ggplot (complete_metrics, aes (x = habitat, y = richness,colour = trap, fill = trap)) + geom_boxplot (width = 0.1 , outlier.shape = NA , alpha = 0.7 , position = position_dodge (0.3 ) ) + geom_point (size = 1.3 , alpha = .3 ,position = position_jitterdodge (dodge.width = 0.85 , jitter.width = 0.1 ,jitter.height = 0.35 )) + xlab ("" ) + ylab ("Richness" ) + scale_colour_manual (values = c ("#440154FF" , "#F9A242FF" ),guide = "none" ) + scale_fill_manual (name = "Trap Type" ,values = c ("#440154FF" , "#F9A242FF" )) + scale_x_discrete (limits = c ("Old-growth Forest" ,"Logged Forest" ,"Riparian Reserve" )) + theme_minimal () + theme (panel.grid.minor.y = element_blank (),panel.grid.major.x = element_blank (),text = element_text (family = "Roboto" ,size = 14 )) + theme (legend.position = "none" )<- ggplot (complete_metrics, aes (x = habitat, y = shannon,colour = trap, fill = trap)) + geom_boxplot (width = 0.1 , outlier.shape = NA , alpha = 0.7 , position = position_dodge (0.3 ) ) + geom_point (size = 1.3 ,alpha = .3 ,position = position_jitterdodge (dodge.width = 0.85 ,jitter.width = 0.1 )) + xlab ("" ) + ylab ("Shannon Index" ) + scale_colour_manual (values = c ("#440154FF" , "#F9A242FF" ),guide = "none" ) + scale_fill_manual (name = "Trap Type" ,values = c ("#440154FF" , "#F9A242FF" )) + scale_x_discrete (limits = c ("Old-growth Forest" ,"Logged Forest" ,"Riparian Reserve" )) + theme_minimal () + theme (panel.grid.minor.y = element_blank (),panel.grid.major.x = element_blank (),text = element_text (family = "Roboto" ,size = 14 )) + theme (legend.position = "none" )<- ggplot (complete_metrics, aes (x = habitat, y = FRic,colour = trap, fill = trap)) + geom_boxplot (width = 0.1 , outlier.shape = NA , alpha = 0.7 , position = position_dodge (0.3 ) ) + geom_point (size = 1.3 ,alpha = .3 ,position = position_jitterdodge (dodge.width = 0.85 ,jitter.width = 0.1 ,jitter.height = 0.3 )) + xlab ("" ) + ylab ("Functional Richness" ) + scale_colour_manual (values = c ("#440154FF" , "#F9A242FF" ),guide = "none" ) + scale_fill_manual (name = "Trap Type" ,values = c ("#440154FF" , "#F9A242FF" )) + scale_x_discrete (limits = c ("Old-growth Forest" ,"Logged Forest" ,"Riparian Reserve" )) + theme_minimal () + theme (panel.grid.minor.y = element_blank (),panel.grid.major.x = element_blank (),text = element_text (family = "Roboto" ,size = 14 )) + theme (legend.position = "none" )<- ggplot (complete_metrics, aes (x = habitat, y = FEve,colour = trap, fill = trap)) + geom_boxplot (width = 0.1 , outlier.shape = NA , alpha = 0.7 , position = position_dodge (0.3 ) ) + geom_point (size = 1.3 ,alpha = .3 ,position = position_jitterdodge (dodge.width = 0.85 ,jitter.width = 0.1 )) + xlab ("" ) + ylab ("Functional Evenness" ) + scale_colour_manual (values = c ("#440154FF" , "#F9A242FF" ),guide = "none" ) + scale_fill_manual (name = "Trap Type" ,values = c ("#440154FF" , "#F9A242FF" )) + scale_x_discrete (limits = c ("Old-growth Forest" ,"Logged Forest" ,"Riparian Reserve" )) + theme_minimal () + theme (panel.grid.minor.y = element_blank (),panel.grid.major.x = element_blank (),text = element_text (family = "Roboto" ,size = 14 )) + theme (legend.position = "none" )<- ggplot (complete_metrics, aes (x = habitat, y = FDis,colour = trap, fill = trap)) + geom_boxplot (width = 0.1 , outlier.shape = NA , alpha = 0.7 , position = position_dodge (0.3 ) ) + geom_point (size = 1.3 ,alpha = .3 ,position = position_jitterdodge (dodge.width = 0.85 ,jitter.width = 0.1 )) + xlab ("" ) + ylab ("Functional Dispersion" ) + scale_colour_manual (values = c ("#440154FF" , "#F9A242FF" ),guide = "none" ) + scale_fill_manual (name = "Trap Type" ,values = c ("#440154FF" , "#F9A242FF" )) + scale_x_discrete (limits = c ("Old-growth Forest" ,"Logged Forest" ,"Riparian Reserve" )) + theme_minimal () + theme (panel.grid.minor.y = element_blank (),panel.grid.major.x = element_blank (),text = element_text (family = "Roboto" ,size = 14 )) + theme (legend.position = "none" )# Patch plots together <- complete_abd_p + complete_rich_p + complete_shan_p + + complete_FEve_p + complete_FDis_p + plot_layout (ncol = 3 , guides = 'collect' ) + plot_annotation (tag_levels = 'A' ) & theme (text = element_text ('Roboto' ))